Saturday, December 27, 2014
The Mix has moved
Stay up to date with the latest stories and insights from UAB research at our new location, www.uab.edu/mix.


Monday, December 22, 2014
Change agent: Creating new scans to track brain diseases
Seven weeks after weight-loss surgery, a group of women have seen significant changes in their body shapes and sizes. They’re each down 20 to 30 pounds, but that’s not the only change their bodies are going through.
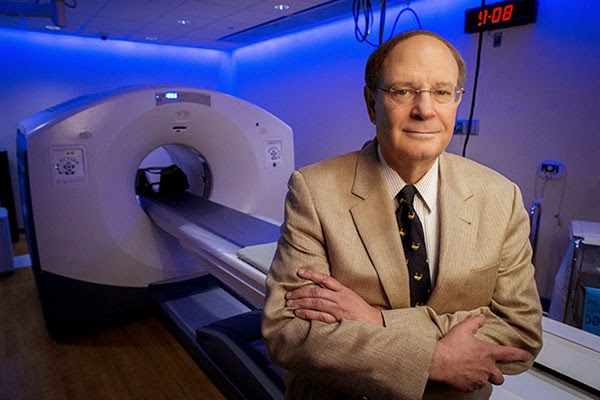
The women’s weight loss is caused by a change in appetite, which results from changes in brain function, explains UAB neuroradiologist Robert Kessler, M.D. (pictured above in UAB's Advanced Imaging Facility). On positron emission tomography (PET) scans, Kessler can see an obvious transformation in the women’s brains, particularly in dopamine neurotransmission.
Using a specialized brain PET scan that he has developed, Kessler can visualize levels of dopamine receptors — molecules that help transmit the brain’s messages of motivation and reward. Before surgery, the women had increased levels of the receptors, which appear on the PET scans as glowing white patches throughout the brain. But after their surgeries, these changes have faded; the women’s brains exhibit a more balanced map of dopamine receptors. In real-world terms, Kessler thinks, these tempered receptor levels reflect a shift to a more normal reward perception, helping the women control their appetites after surgery.
Obesity — and the drive to overeat — isn’t the only pathology that Kessler can see when he peers into people’s brains with a PET scan. During the past 30 years, he has helped illuminate changes to the brain that might underlie schizophrenia, drug addiction, depression and dementia, among other disorders. By looking at a person’s brain PET scan and carefully measuring the levels of neurotransmitter function, Kessler can tell whether someone is more prone to taking risks than average, whether they’re more of a “slacker” or a “go-getter,” and whether or not they have “the ability to experience rewarding stimuli in a normal manner or if they have lost that ability,” he said.
Kessler, who joined the UAB faculty in 2013 as director of neurochemical brain imaging and PET neurotracer development in the Department of Radiology, says these specialized PET scans are paving the way toward a new level of understanding of brain diseases.
“At a very basic scientific level, there’s no other technology that can look at the human brain and inform you about specific molecules and receptors,” Kessler said. At UAB, he’s taking advantage of the university’s TR24 cyclotron — the largest at any U.S. academic medical center — to develop new PET scans. And he has launched collaborations with UAB researchers across the psychiatric and neurological sciences to help them apply his techniques to even more questions.
In 1977, Kessler joined a lab at the National Institutes of Health just as this was changing. One of his mentors there became the first to use a PET scan to visualize the activity of the brain. The earliest scans, rather than pinpointing specific receptors as Kessler does now, were designed to simply show which cells in the brain were undergoing metabolism — a sign of activity — at any given moment.
But the basic idea has been the same for more than three decades now: A patient gets an injection of a radioactive tracer into their bloodstream. Depending on the design of the tracer, it accumulates in particular organs or cells of the body. Then, a PET machine is used to measure the location of the accumulated radioactivity.
“It quickly became clear to me that PET was going to become an important tool for understanding the brain,” Kessler said. “And we began to use it to look at everything from brain tumors and schizophrenia to aging and dementia.”
As Kessler immersed himself in the new technology, first at NIH and then at Vanderbilt University, he helped develop new tracers that would pave the way for the rest of his career: 18F-Fallypride, and later 18F-FPEB. Rather than building up in all metabolizing brain cells, these radioactive molecules bind specifically to dopamine and glutamate receptors.
Among the first questions Kessler asked with 18F-Fallypride was ‘What are the effects of antipsychotic drugs on the brains of patients with schizophrenia?’ A new class of antipsychotic drug had recently been developed; the drugs had fewer side-effects than older versions, but researchers didn’t know why. Kessler and his collaborators discovered that the new drugs targeted different areas of the brain than the old drugs, offering not only an explanation for the differences, but a way to test future drugs for their efficacy.
Dopamine had also already become known as a chemical that mediates reward-seeking behavior and pleasure. So Kessler’s lab began to look at how levels of the dopamine receptor and the effects of dopamine release on dopamine receptors might relate to drug abuse, impulse control, addiction and the ability to feel pleasure.
“People who are depressed lose their ability to enjoy rewards and experience the pleasures of life; people who are addicted have very distorted reward functions where they crave just one reward,” Kessler said. “We showed that dopamine plays a key role in all of these.”
One of the ongoing challenges in the field, Kessler says, is developing new tracers. With UAB’s new cyclotron, Kessler says he has the tools at his fingertips to continue developing and perfecting tracers that bind to different receptors in the brain. He’s already begun work with the neurotransmitter glutamate; like dopamine, glutamate can be studied through PET tracers that bind to glutamate receptors. And glutamate is thought to have roles in autism, Huntington’s disease, Parkinson’s and anxiety disorders, among other things.
This fall, Kessler launched a study examining glutamate receptors in the brains of addicts as they voluntarily withdraw from methamphetamine. The findings — if they show key differences from normal brains — could lead to new drugs to help meth addicts quit their addiction. Studies on Parkinson’s, depression and Alzheimer’s disease are also in the works with UAB collaborators.
Although “this is a tough area to work in for many reasons,” Kessler said — from the vagaries of chemical half-lives to the sheer complexity of the brain itself — he wouldn’t have it any other way. “You just can’t get this kind of information anywhere else.”
The women’s weight loss is caused by a change in appetite, which results from changes in brain function, explains UAB neuroradiologist Robert Kessler, M.D. (pictured above in UAB's Advanced Imaging Facility). On positron emission tomography (PET) scans, Kessler can see an obvious transformation in the women’s brains, particularly in dopamine neurotransmission.
Using a specialized brain PET scan that he has developed, Kessler can visualize levels of dopamine receptors — molecules that help transmit the brain’s messages of motivation and reward. Before surgery, the women had increased levels of the receptors, which appear on the PET scans as glowing white patches throughout the brain. But after their surgeries, these changes have faded; the women’s brains exhibit a more balanced map of dopamine receptors. In real-world terms, Kessler thinks, these tempered receptor levels reflect a shift to a more normal reward perception, helping the women control their appetites after surgery.
Obesity — and the drive to overeat — isn’t the only pathology that Kessler can see when he peers into people’s brains with a PET scan. During the past 30 years, he has helped illuminate changes to the brain that might underlie schizophrenia, drug addiction, depression and dementia, among other disorders. By looking at a person’s brain PET scan and carefully measuring the levels of neurotransmitter function, Kessler can tell whether someone is more prone to taking risks than average, whether they’re more of a “slacker” or a “go-getter,” and whether or not they have “the ability to experience rewarding stimuli in a normal manner or if they have lost that ability,” he said.
| Find out how a cyclotron works, and what makes UAB’s new cyclotron unique among U.S. academic medical centers, in the video above and in this feature from UAB Magazine. |
“At a very basic scientific level, there’s no other technology that can look at the human brain and inform you about specific molecules and receptors,” Kessler said. At UAB, he’s taking advantage of the university’s TR24 cyclotron — the largest at any U.S. academic medical center — to develop new PET scans. And he has launched collaborations with UAB researchers across the psychiatric and neurological sciences to help them apply his techniques to even more questions.
Focusing on Receptors
As a medical student, resident and fellow in the 1970s, Kessler first became interested in the human brain at a time that clinicians had few methods to visualize the organ. Surgeons could physically see the outer layers of the brain when they opened the skull for an operation, or pathologists could dissect an autopsied brain; but viewing the activity — in a living human — of the molecules that make up the brain’s electrical pathways wasn’t possible.In 1977, Kessler joined a lab at the National Institutes of Health just as this was changing. One of his mentors there became the first to use a PET scan to visualize the activity of the brain. The earliest scans, rather than pinpointing specific receptors as Kessler does now, were designed to simply show which cells in the brain were undergoing metabolism — a sign of activity — at any given moment.
But the basic idea has been the same for more than three decades now: A patient gets an injection of a radioactive tracer into their bloodstream. Depending on the design of the tracer, it accumulates in particular organs or cells of the body. Then, a PET machine is used to measure the location of the accumulated radioactivity.
“It quickly became clear to me that PET was going to become an important tool for understanding the brain,” Kessler said. “And we began to use it to look at everything from brain tumors and schizophrenia to aging and dementia.”
As Kessler immersed himself in the new technology, first at NIH and then at Vanderbilt University, he helped develop new tracers that would pave the way for the rest of his career: 18F-Fallypride, and later 18F-FPEB. Rather than building up in all metabolizing brain cells, these radioactive molecules bind specifically to dopamine and glutamate receptors.
Among the first questions Kessler asked with 18F-Fallypride was ‘What are the effects of antipsychotic drugs on the brains of patients with schizophrenia?’ A new class of antipsychotic drug had recently been developed; the drugs had fewer side-effects than older versions, but researchers didn’t know why. Kessler and his collaborators discovered that the new drugs targeted different areas of the brain than the old drugs, offering not only an explanation for the differences, but a way to test future drugs for their efficacy.
Dopamine had also already become known as a chemical that mediates reward-seeking behavior and pleasure. So Kessler’s lab began to look at how levels of the dopamine receptor and the effects of dopamine release on dopamine receptors might relate to drug abuse, impulse control, addiction and the ability to feel pleasure.
“People who are depressed lose their ability to enjoy rewards and experience the pleasures of life; people who are addicted have very distorted reward functions where they crave just one reward,” Kessler said. “We showed that dopamine plays a key role in all of these.”
 |
| These are 18F-Fallypride PET images of dopamine D2 type receptors, averaged across several normal subjects. There are high levels of these receptors (red color) in deep brain structures and lower levels in the cortex. These include the basal ganglia and thalamus (A), amygdala and temporal cortex (B), and substantia nigra (C). These regions are concerned with movement, emotion and cognition. |
The Future of Brain PET
PET scans offer the most direct way to observe what happens at a molecular level in the brain when someone develops, or recovers from, a psychiatric disorder or addiction, Kessler says. Drug developers and pharmaceutical companies now use PET scans to fine-tune prospective new treatments, he notes. If they know they need to lower the number of dopamine receptors in one area of the brain, for instance, they can use PET scans to determine which drugs, and drug dosages, effectively achieve this.One of the ongoing challenges in the field, Kessler says, is developing new tracers. With UAB’s new cyclotron, Kessler says he has the tools at his fingertips to continue developing and perfecting tracers that bind to different receptors in the brain. He’s already begun work with the neurotransmitter glutamate; like dopamine, glutamate can be studied through PET tracers that bind to glutamate receptors. And glutamate is thought to have roles in autism, Huntington’s disease, Parkinson’s and anxiety disorders, among other things.
This fall, Kessler launched a study examining glutamate receptors in the brains of addicts as they voluntarily withdraw from methamphetamine. The findings — if they show key differences from normal brains — could lead to new drugs to help meth addicts quit their addiction. Studies on Parkinson’s, depression and Alzheimer’s disease are also in the works with UAB collaborators.
Although “this is a tough area to work in for many reasons,” Kessler said — from the vagaries of chemical half-lives to the sheer complexity of the brain itself — he wouldn’t have it any other way. “You just can’t get this kind of information anywhere else.”
Monday, December 15, 2014
Equations against cancer: Using math to predict a tumor's path
Hassan Fathallah-Shaykh, M.D., Ph.D., believes that math can transform medicine, and he has the numbers to prove it.
In the clinic, this UAB neurologist specializes in treating brain tumors. In his lab at the Comprehensive Cancer Center, Fathallah-Shaykh, who is also a professor of mathematics at UAB, wields equations as well as petri dishes. His mathematical models of cancer behavior are offering new insights on tumor growth. Eventually, they could be used to personalize treatment based on the unique characteristics of each patient’s cancer cells and anatomy.
Fathallah-Shaykh is one of a growing number of researchers worldwide exploring the field of mathematical biology, which “uses mathematical tools to generate models of biological problems,” he said. Building mathematical models based on the current understanding of a disease, for example, allows researchers to “test whether the assumptions are accurate,” Fathallah-Shaykh said.
 |
| Hassan Fathallah-Shaykh |
Model Behavior
Working with colleagues at the University of Bordeaux, and UAB graduate student Elizabeth Scribner, Fathallah-Shaykh has created an elegant model of the aggressive brain cancer glioblastoma multiforme (GBM). It produces simulations on the scale of clinical MRI scans, so that its predictions can be tested directly against patient data. In a paper published on Dec. 15 in PLOS ONE, the researchers demonstrated that their model can reproduce the typical GBM growth patterns seen on patient scans. They also revealed its value as a research tool.The model predicted a previously unknown pattern of tumor growth in patients with recurrent GBM treated with the anti-angiogenesis drug bevacizumab. This growth, powered by a cycle of proliferation and brain invasion, is characterized by an expanding area of invasive cells and dead cells known as necrosis, the researchers say. A subsequent search of 70 patient MRI scans by the researchers turned up the same pattern in 11 cases.
“We hope to tailor radiation therapy, chemotherapy and other treatments based on a personalized model of a patient’s tumor.”
That pattern explains the disappointing results of recent Phase III clinical trials of anti-angiogenesis therapies against GBM, the researchers say. Anti-angiogenesis drugs attempt to kill tumors by preventing them from growing new blood vessels. But the model demonstrated how GBM cells can flee from the oxygen-depleted treatment area — and quickly begin expanding again as soon as therapy stops or the tumor becomes resistant to the drugs. (For more on the model and these findings, see “SimTumor,” below.)
“We’ve shown that we can predict new insights on cancer behavior,” Fathallah-Shaykh said. The results have already spurred Fathallah-Shaykh to pursue new therapies in his lab to limit tumor mobility. Ultimately, the researchers hope to use their model to personalize therapy to the unique characteristics of a patient’s tumor. They could do that by analyzing the existing growth pattern of a tumor and building that into the model’s parameters. Then they could simulate its future behavior on a virtual MRI slice that reproduces the unique anatomy of the patient’s brain. “We hope to tailor radiation therapy, chemotherapy and other treatments based on a personalized model of a patient’s tumor,” said Fathallah-Shaykh.
Advancing Mathematical Biology ResearchThis spring, Fathallah-Shaykh helped organize a symposium on the topic as part of the College of Arts and Sciences’ Interdisciplinary Innovation Forum series. The meeting attracted some of the mathematical biology’s most famous names. Meanwhile, he is helping to attract new talent to the discipline by teaching undergraduate and graduate courses on Mathematical Biology in the math department. “It is quite clear that the next great advances in medicine cannot happen without math,” Fathallah-Shaykh said. “These are exciting times.” |
From Flies to Colon Cancer
Since he joined the UAB faculty in 2008, Fathallah-Shaykh has been developing ever more advanced models to predict the behavior of biological networks. He began by building a model of the molecular clock in a fruit fly’s brain. Despite the fly’s tiny size, it’s a challenging puzzle. The clock is a tangled web of positive and negative feedback loops, with five different genes producing proteins that inhibit and activate one another (as well as themselves, in some cases) in a regular cycle.First, Fathallah-Shaykh and his collaborators “showed we can replicate everything the clock is known to do,” he said. Then they proved it was a useful research tool, answering a perplexing question about the fruit-fly gene Clockwork Orange that had stumped biologists for years.
The researchers next adapted their model to track the developing neural networks in fruit-fly embryos. To do this, they utilized the Kalman filter, a mathematical technique to analyze and predict changes that helps track planes in flight. Now, “we’re using the model to study molecular networks in colon cancer,” Fathallah-Shaykh said.
Coping with an Information Explosion
Fathallah-Shaykh has always been fascinated with math. “It’s like a symphony; it’s beautiful,” he said. “But it’s also very applicable.” He cemented the connection between medicine and math as a neurologist at Rush University Medical Center in Chicago when he enrolled in a doctoral program in mathematics at the nearby University of Illinois–Chicago. “I would go to class in between patients,” he said.Math is essential to making progress against the toughest questions in medicine, Fathallah-Shaykh contends. To illustrate the problems that researchers face, he points to a chart of all the known molecular pathways involved in Alzheimer’s disease. It’s a mass of interlocking loops and tangles that fills an entire page. Researchers specialize in tiny sections of this wiring diagram, but understanding how it all works together is another problem entirely. Even worse, these networks are intertwined in such a way that multiple paths can lead to the same destination. That may help explain why treatments that work beautifully in isolated cell lines in a lab so often fail when they encounter the complex networks of the body.
There’s another wrinkle. “Cells migrate, they communicate, they interact with one another over time,” said Fathallah-Shaykh. The waves of mutations, which are a hallmark of cancer, make the problem particularly complex, he noted. “Whole pathways are deleted and new connections start turning up.” It’s a perfect example of a nonlinear dynamic system, like the weather or the stock market, in which slight changes in one parameter can lead to wildly diverging outcomes.
The good news, said Fathallah-Shaykh, is that “mathematics has very rich tools” to model just these types of systems, as he has demonstrated with his cancer simulations. But this work has another exciting element for Fathallah-Shaykh as a mathematician: It opens new horizons in math theory. “Equations have already been developed from biological problems,” he said, “and there is very strong evidence that they will produce spectacular advances in mathematics.”
SimTumor
At the heart of Hassan Fathallah-Shaykh’s new mathematical model of glioblastoma multiforme (GBM) are 10 partial differential equations. Here’s how it works — and what it has revealed about GBM behavior.
Formula 10
Equations track each of four different cell types, with unique rules of behavior.
Proliferative GBM cells (P), which make up the bulk of the tumor, divide but don’t move.
Invasive GBM cells (I), found on the fringes of the tumor, move but don’t divide.
Healthy brain cells (B) neither divide nor move, although they are displaced by the growing tumor.
Cells in the center of the tumor, cut off from nourishing blood vessels, are starved of oxygen (hypoxia) and die, becoming necrotic cells (N).
The remaining six equations track angiogenesis (new blood vessel formation), oxygen levels, and rates of necrosis and cell division.
Built for Speed
Fathallah-Shaykh’s first GBM model, published in August 2014 in the Bulletin of Mathematical Biology, consisted of many more equations. It required a supercomputer, and several days, to run. The model published in PLOS ONE can run in 50 seconds on a typical desktop computer.
And They’re Off!
The simulation begins with a tiny clump of tumor cells surrounded by healthy brain. As the program continues over several virtual weeks, this mass expands in the characteristic manner seen on patient MRIs, with a dark region of necrotic cells in the center, surrounded by a large group of proliferative cells and an outer rim of invasive cells.
Grow or Go
The model’s main assumption is that proliferative cells can turn into invasive cells in hypoxic conditions. This is in keeping with the “grow or go” hypothesis of GBM behavior, which says that low oxygen levels spur GBM cells to flee the dying core of the tumor. When these new invasive cells reach healthy, oxygenated areas of brain, they switch back into proliferative mode and start growing again.
How GBM Escapes Anti-Angiogenesis Therapy
As tumors grow, cells at the core lose contact with nourishing blood vessels and die.
To get around this problem, tumors release VEGF (vascular endothelial growth factor), which induces the body to create new blood vessels (a process known as angiogenesis). In fact, the well-known Folkman Hypothesis states that tumors must be able to induce blood vessel growth in order to keep growing.
Clinicians had high hopes that anti-angiogenesis medications such as bevacizumab (Avastin), could keep tumor growth in check. But two high-profile Phase III clinical trials, which released results in early 2014, found that bevacizumab therapy did not prolong overall survival in patients with recurrent GBM, although it did extend progression-free survival and patient quality of life.
Fathallah-Shaykh’s model, programmed to simulate the effects of anti-angiogenesis therapy, reveals an explanation for this “unusual clinical finding.” When bevacizumab therapy causes oxygen levels to drop, proliferative cells turn into invasive cells and flee the scene. When they reach an area with sufficient oxygen, they convert back into proliferative cells and begin a new cycle of growth. This sets up the tumor for rapid “rebound” growth as soon as it becomes resistant to bevacizumab or therapy is discontinued. That explains why patients treated with bevacizumab in the recent trials didn’t experience any increase in overall survival rates over those who were not treated.
Toward New Treatment Approaches
The model underlines the importance of better understanding the molecular mechanisms of brain cell invasion, particularly the active transport of invasive cells toward healthy brain regions, says Fathallah-Shaykh.
There are currently no available biomarkers to identify the quantity of invasive cells in a patient’s tumor. But finding such a biomarker, and drugs that can target these cells to prevent tumor migration, is a current research focus in Fathallah-Shaykh’s lab. “If we’re going to kill these tumors,” he said, “we have to target the cells that are invading.”
Tuesday, December 2, 2014
Creating a roadmap to bring innovative medical technologies to market
Robert Hergenrother, Ph.D., isn’t a surgeon, but he has done some of his best work in the operating room. In his three decades in the medical device industry, Hergenrother has led engineering teams that have created 15 products, including new technologies for use in brain surgery, wound care and diagnosing disease. “When you have a surgeon come up to you and say, ‘If it wasn’t for your device, I couldn’t have helped that patient,’ that’s pretty powerful,” Hergenrother said.
As the director of the new Alliance for Innovative Medical Technologies (AIMTech), Hergenrother is focused on creating the next generation of life-changing medical devices in Birmingham. AIMTech is a partnership between UAB and Southern Research Institute, modeled after the two institutions’ successful Alabama Drug Discovery Alliance. It will identify promising projects already in development at both institutions and launch new projects that meet pressing clinical needs, Hergenrother explains. AIMTech will provide investment and support to bring these projects through clinical trials and FDA approval. Then the devices will be spun off in startup companies or licensed to major medical device makers.
Scouting for the Next Big Thing
Hergenrother, who also has a faculty appointment in the UAB Department of Biomedical Engineering, is now meeting with clinicians and researchers across campus. “In one day I can go from drug delivery to sports medicine to physical therapy to radiology,” he said. “I get to see a lot of different ideas and work with people who really are excited about moving these ideas forward.”UAB Department of Physical Therapy. Others are simply intriguing concepts. Hergenrother recently met with a surgeon who wants to develop a new tool for cartilage repair. “He had an idea and a drawing,” Hergenrother said. “We were able to come in and make a quick prototype and put it in his hands. He was fired up. It’s a great way to just start answering questions: ‘Is this working, yes or no?’”
Conversations with UAB clinicians will lead to opportunities to create entirely new types of devices. “We want to focus on what is causing people problems now,” Hergenrother said. One of his main jobs, he explains, is to connect clinicians with researchers who can develop solutions to meet their needs.
Hergenrother, who holds 18 patents of his own, understands the thrill of a new invention. But creating a successful medical device isn’t a matter of innovation alone, he points out. As part of the initial scouting phase of the program, “I’m asking investigators to work with me to conduct 20 interviews with the people who will be the ultimate end-users of their product,” he said. “They need to find out how people are doing the job now, what the current solutions are and what advantages their product has to offer.”
Competitive Advantages
Major medical device companies are always eager for new ideas, Hergenrother says. As the industry matures — it is projected to grow by nearly 21 percent by 2016 — those companies are focusing more on international expansion and production efficiencies, he adds. “They’re relying on smaller companies and universities to drive innovation.”Top TargetsAIMTech will initially focus on developing projects in five key areas: | |
| Cardiology |  |
| Orthopedics |  |
| Ophthalmology |  |
| Rehabilitation Engineering |  |
| Trauma |  |
In industry jargon, this is known as “de-risking” — building up the scientific and marketing data necessary to justify a major financial investment. AIMTech will be able to supply that proof by tapping into the combined capabilities of Southern Research and UAB.
UAB’s Institute for Innovation and Entrepreneurship (IIE) will vet the intellectual property position of every project that enters the AIMTech program. IIE and Southern Research will also evaluate potential market size, regulatory pathways and reimbursement strategies so that only the strongest, most market-ready technologies advance through AIMTech’s pipeline. Southern Research has extensive experience in assembling product-development systems and negotiating the regulatory requirements of clinical trials and FDA approvals, Hergenrother notes. “Someone has to do that work,” he said. “If we can take our projects further along than another university, ours will be more attractive to potential partners.”
AIMTech’s ultimate mission is to get life-changing products to market as quickly as possible, Hergenrother says. “We always want to keep in mind why we’re doing this. It’s not to get another patent, but to save lives. We have an opportunity to really make a difference here.”
Tuesday, November 18, 2014
Immunogenomics advances point to new biomarkers, therapies
Next-generation gene-sequencing technology and new data-analysis tools are pointing the way to fresh diagnostic and treatment approaches for autoimmune diseases, cancer and many other conditions. That was the message at Immunogenomics 2014, a recent conference hosted by Huntsville’s HudsonAlpha Institute for Biotechnology and Science magazine for researchers studying the interaction between genes and the immune system. The event was sponsored in partnership with UAB and its Comprehensive Arthritis, Musculoskeletal and Autoimmunity Center (CAMAC).
Investigators from major national and international research institutions described how detailed profiles of immune cells could improve response to influenza vaccines and accelerate new treatments for emerging infectious diseases. They explained new immune-mediated links between microbial populations and cancer risk, and highlighted progress in understanding the pathogenesis of complex diseases such as multiple sclerosis.
“We now have the tools to examine these gene-disease associations in finer detail,” said S. Louis Bridges Jr., M.D., Ph.D., director of UAB’s Division of Clinical Immunology and Rheumatology and the CAMAC. Bridges is using genomic techniques to study the autoimmune condition rheumatoid arthritis. Bridges presented his research at Immunogenomics 2014, and served as chair of a session on the genetics of complex disease. “We’ve started to refine our analysis to home in on cells with an increasing degree of specificity,” Bridges said. “Technology is now allowing us to analyze in more detail specific subsets of cells, including ultimately at the single-cell level.”
From Associations to Biomarkers
Genomewide association studies have identified a host of genetic changes linked with disease. “Now investigators are looking at the functional effects of these polymorphisms,” Bridges said. “It could be that a polymorphism affects expression of a certain gene, or it may only affect expression of that gene in a certain cell type.” Bridges’ project, being performed in collaboration with UAB epidemiology chair Donna Arnett, Ph.D., and HudsonAlpha investigator Devin Absher, Ph.D., is examining genetic risk factors in African-Americans with RA."We now have the tools to examine these gene-disease associations in finer detail," Bridges said. "We've started to refine our analysis to home in on cells with an increasing degree of specificity."
In his talk at Immunogenomics 2014, Bridges demonstrated that overexpression of the interferon gamma 2 receptor gene is strongly linked with severity of disease in African-American patients with RA. “Our next step is to see in which cells that particular expression occurs,” Bridges said. This work could ultimately point the way to biomarkers that tell clinicians which of several known signaling pathways is active in a patient with RA, guiding treatment decisions.
Epigenetics and Therapeutics
Researchers are also focusing increasing attention on the ways gene expression is regulated dynamically in cells through epigenetic changes, says Robert P. Kimberly, M.D., director of the UAB Center for Clinical and Translational Science. Epigenetics refers to mechanisms that alter gene expression without changes in the actual DNA sequence. One of the most common epigenetic changes is methylation. When a methyl group attaches to the DNA base cytosine, it blocks the ability of the neighboring gene to be expressed.Tracking and analyzing epigenetic markers implicated in a particular disease — such as systemic lupus erythematosus, one of Kimberly’s own research interests — could give clinicians crucial information on “if to treat, when to treat and also how to treat” that condition, he said.
For example, if a key risk gene is hypo-methylated in a patient — increasing the likelihood of the gene’s being expressed — “that could mean the patient is poised for a flare-up of disease,” Kimberly said. Another patient, who would look exactly the same to a clinician, would be much less likely to have a flare-up if that gene is hyper-methylated, he adds. “Understanding this epigenetic regulation, and eventually manipulating it to therapeutic advantage, is very exciting.” This research is a focus of several investigative teams at UAB, Kimberly says.
Profiling Immune Signatures
One advance highlighted by several presenters at Immunogenomics 2014 was immune repertoire sequencing. Researchers now understand that an individual’s immune response is based greatly on the specific cell populations, or “repertoire,” present in that individual. For both T and B cells, for example, millions of distinct variants are possible. The exact mix is determined by a person’s encounters with microbes, disease and other environmental exposures over a lifetime, explains HudsonAlpha investigator Jian Han, M.D., Ph.D., a 1991 graduate of UAB’s medical genetics doctoral program.Everyone produces T cells, for example, Han says. “But which one of those naive T cells gets used is determined by if it met, and was activated by, its antigen.” By analyzing the variety of T and B cells present in a particular patient, or mapping the repertoire commonly found in a particular disease, researchers can identify biomarkers to aid in diagnosis and treatment, Han notes.
Han’s HudsonAlpha lab has pioneered the multiplex PCR technology needed to gather the massive amounts of data required for repertoire sequencing, and the analytical tools required to resolve that data into meaningful reports. His presentation at Immunogenomics 2014 focused on Repertoire10K, a HudsonAlpha-funded project to sequence the immune repertoires of 10,000 patients: 100 with each of 100 critical diseases. The goal is to identify a signature in the immune repertoire for each disease. UAB researchers have been key contributors of the genetic samples that are critical to the project, Han says. In return, the investigators have access to state-of-the-art sequencing data that can advance their own studies.
Team-based Science
Collaborations between investigators at HudsonAlpha and UAB have taken place since the institute first opened in 2008, Kimberly says. But the new UAB–HudsonAlpha Center for Genomic Medicine, launched this summer, will increase these research partnerships and speed new discoveries in immunology, cancer, cardiovascular disease and many other fields, he notes.Leveraging the strengths of each institution is critical as the scale of the research challenges becomes ever greater, Kimberly adds. “To understand what’s really happening in disease states, we’re going to have to be able to take all the data on genomics, epigenetics and more and figure out how to pull it all together,” he said.
That’s why UAB is also creating a new Informatics Institute. It will work in tandem with the UAB–HudsonAlpha Center for Genomic Medicine and a third initiative, the UAB Personalized Medicine Institute, to build the infrastructure and recruit the data scientists needed to succeed in this new era of research.
“It’s a major frontier right now,” Kimberly said. “The algorithms to combine all this data for the most part haven’t even been formulated yet. But it’s clear that the institutions that succeed in the future will be the innovators in this area.”
Labels:
autoimmunity,
genomics,
HudsonAlpha,
immunogenomics,
immunology
Thursday, November 6, 2014
Discovery route: Path to potential diabetes drugs began with a simple question
 |
| More than 12 years of research led Anath Shalev, M.D. (right, with Junqin Chen, Ph.D.) from a basic discovery to the first human trial of a new type of diabetes drug. |
In 2002, diabetes researcher Anath Shalev, M.D., asked a basic question: What gene in the insulin-producing islets of the human pancreas is most turned on by high levels of glucose, a hallmark of diabetes?
The answer has led the UAB endocrinologist to discover new cellular pathways in beta cells of the islets, pathways that are a key to diabetes progression or protection. Those discoveries have now opened the door to the first human trial of a potential diabetes drug with a mode of action different from any current diabetes treatment. (Learn more about the trial, which will begin in early 2015, in this story.)
| Anath Shalev explains verapamil's protective effects against diabetes, and a new human clinical trial of the drug at UAB, in this video. |
From Molecular Mechanisms to New Treatments
The beta-cell gene that responded to the high glucose in Shalev’s 2002 experiment produces TXNIP (pronounced "ticks-nip"), a protein normally involved in controlling oxygen radicals in many types of cells but never known to be important in beta-cell biology. Its response to glucose was intriguing because TXNIP (thioredoxin-interacting protein) was already recognized as a regulator of thioredoxin. Overexpression of thioredoxin had previously been shown to prevent experimentally induced diabetes by inhibiting the programmed death of islet beta cells. Since TXNIP inhibits thioredoxin, and because Shalev had discovered that islet TXNIP was highly regulated by glucose, Shalev realized that TXNIP might have major implications for beta-cell biology.What does it take to go from a basic microarray gene discovery to a human trial of a completely novel drug to treat diabetes?
A dozen years of elegant research unraveling the control and function of a protein called TXNIP.
Over the next dozen years, Shalev — who left the University of Wisconsin–Madison to head the UAB Comprehensive Diabetes Center in 2010 — set out to reveal how TXNIP acts in cells at the molecular level, knowing that an understanding of those molecular mechanisms might point to possible new diabetes treatments. The payoff has been substantial: Using cell cultures, mouse models and pancreatic islets isolated from humans, the Shalev lab team has shown that manipulating TXNIP levels up or down in beta cells could exacerbate or protect against experimental diabetes.
Details about the research journey show the incremental steps that basic science takes, and how those connected steps sometimes lead to potential clinical impacts.
Controlling TXNIP to Treat Diabetes
In 2005, the Shalev lab team found that beta-cell TXNIP levels are higher in mouse diabetes models, and that experimentally increasing TXNIP levels in rat beta cells in vitro led to increased programmed cell death, by means of a well-known trigger signal of apoptosis. The Shalev team also found that sugars in general, whether metabolized or not, turn the TXNIP gene on. This clue led them to a newly identified carbohydrate response element (ChoRE) in the TXNIP promoter that acts as a regulator of TXNIP.
In 2008, the Shalev lab developed mice that had little or no TXNIP in their beta cells. These lower levels protected against experimental diabetes. The team also discovered that the lower levels sent a known signal that inhibited mitochondrial beta-cell death. Shalev wrote, “These results suggest that lowering beta-cell TXNIP production could serve as a novel strategy for the treatment of type 1 and type 2 diabetes by promoting endogenous beta-cell survival.”
An Approved Drug Offers Protection
In 2012, the Shalev group tested an already approved oral drug that they had earlier found to reduce levels of TXNIP in heart cells. The drug — verapamil — is a calcium channel blocker used primarily to treat high blood pressure, but also to treat migraine headaches. Shalev’s team found that exposing in vitro beta cells or isolated human islets to verapamil reduced TXNIP expression, and halted programmed apoptotic death of beta cells. Furthermore, mice that were fed verapamil in their drinking water were protected from experimentally induced diabetes, and verapamil rescued mice that already had diabetes. The verapamil mice had lower TXNIP levels and less programmed beta-cell death, as well as better levels of insulin
"I actually went down to the mouse house to see if the mice were getting diabetes," Shalev told The Birmingham News in 2012. When she found normal glucose levels, "We were dancing."
In those studies, the group also revealed how verapamil lowers TXNIP — the decreased intracellular level of calcium ions caused by verapamil led to phosphorylation of the ChoRE binding protein that normally responds to glucose to control TXNIP transcription at the ChoRE. This phosphorylation prevented the binding protein from entering the beta-cell nucleus and interacting with the TXNIP gene. Shalev noted that these verapamil results identified, for the first time, “… an effective pharmacological means … to inhibit pancreatic beta-cell expression of proapoptotic TXNIP, enhance beta-cell survival and function, and thereby prevent and even improve overt diabetes and shed light on the mechanisms involved.”
Another Role for TXNIP, Another Drug Target?
In 2013, TXNIP was shown to play another crucial role in beta-cell biology when the Shalev laboratory team discovered that high levels of TXNIP directly blocked insulin production in beta cells, acting through a newly identified pathway. TXNIP, they found, induced a microRNA called miR-204, which in turn down-regulated the MAFA transcription factor involved in promoting transcription of the insulin gene.
This means that miR-204 may offer another target for a future RNA drug, an area that is currently also being actively pursued by the Shalev lab. MicroRNAs, with 20 to 24 noncoding nucleotides, have rapidly gained prominence as regulators of gene expression in health and disease. Researchers are beginning to explore whether silencing targeted microRNAs may lead to a treatment for cancers or other diseases.
TXNIP's Vicious Cycle
This year Shalev reported that TXNIP — surprisingly — can induce its own transcription. Her UAB research team found that TXNIP does this by affecting the same ChoRE binding protein (ChREBP) that was previously found to be key in the response to the drug verapamil. The researchers experimentally elevated TXNIP levels in beta cells and found this caused decreased phosphorylation of ChREBP, which led to its increased entry into the nucleus and its increased binding to the TXNIP promoter to boost transcription. This creates a harmful positive-feedback loop.
"These findings support the notion,” Shalev wrote in this 2014 paper, “that TXNIP levels rise over time, not only as a result of elevated blood glucose levels and/or endoplasmic reticulum stress, but also as part of a vicious cycle by which increased TXNIP levels lead to more TXNIP expression and thereby amplify the associated detrimental effects on beta-cell biology including oxidative stress, inflammation, and ultimately beta-cell death and disease progression.”
First Human Trial
| Get a quick overview of the science behind UAB's verapamil trial in this animation |
Meanwhile, a UAB partnership with the Southern Research Institute — called the Alabama Drug Discovery Alliance — is already working to develop small therapeutic molecules that mimic the diabetes-protecting effect produced by verapamil and inhibit TXNIP, but have a greater selectivity and efficacy. [Learn more about this work, and other high-potential projects in the Alabama Drug Discovery Alliance, in a new feature from UAB Magazine.]
So Shalev’s simple question — what gene in insulin-producing beta cells is most turned on by glucose? — has thus led the research out of her laboratory to possible new drugs, acting against a novel target to alleviate or reverse diabetes.
— Jeff Hansen
Tuesday, October 28, 2014
Exploring new frontiers in personalized cancer care
Personalized medicine is turning medical care on its head, and cancer treatment is at the forefront of that revolution. The UAB Comprehensive Cancer Center’s 17th Annual Research Retreat introduced this cutting-edge work to an audience of nearly 400 clinicians and researchers. The topic was timely after this summer’s announcement of major initiatives in genomics and personalized medicine at UAB, including a research consortium between the Cancer Center and Huntsville’s HudsonAlpha Institute for Biotechnology.
“Personalized medicine is the future of cancer care,” noted Eddy Yang, M.D., Ph.D., associate professor in the UAB Department of Radiation Oncology, who organized this year’s symposium. “This is certainly a glimpse of what is to come for the Cancer Center and UAB as a whole.”
The Future: Cancer as a Chronic Disease
“Oncology has been a first mover for personalized medicine,” said invited speaker Mark Boguski, M.D., Ph.D., founder of Genome Health Solutions and a faculty member at Harvard Medical School.Boguski shared his remarkable vision. With the use of personalized medicine, he said, we can now begin to reimagine cancer as a manageable chronic disease. Subsequent speakers amplified that theme, describing advances, challenges and roadblocks to delivering personalized cancer care to patients across the United States.
Boguski began with three patient case histories.
The first was a patient in 2010 with adenocarcinoma that was EGFR-positive (that is, it contained mutations that activated the EGFR pathway). When treatment with the usual drug failed, genomic and transcriptomic analysis showed why — metastases from the original cancer were no longer EGFR-positive. But biomarkers on those cancer cells successfully identified a target for a different drug that was effective.
The second case was a metastatic squamous cell carcinoma. Genomic analysis showed, surprisingly, that it could be treated with a hematological cancer drug.
“You wouldn’t guess to use that on a solid tumor,” Boguski said.
Similarly, in a case of advanced lymphoblastic leukemia, genomic analysis unexpectedly pointed to using a renal cell carcinoma drug. With this sea change in the way that oncologists can make their treatment decisions, cancer patients are beginning to ask that their genomes be analyzed, Boguski said.
The UAB Cancer Center’s Molecular Tumor Board, initiated last year, identifies patients who could benefit from DNA sequencing of their tumors, said Yang. These tests, usually conducted in patients with rare tumors or tumors that do not respond to typical treatment, can identify off-label uses for cancer drugs. For example, BRAF inhibitors, which are approved for melanoma, have been used to treat patients with other tumor types that nevertheless harbor the BRAF V600E mutation, Yang said. In another important consideration, “treating physicians have been successful in getting third-party payers to pay for these drugs outside the ‘approved’ indications using the profiling results,” he explained.
Cancer Center Honors Research ExcellenceIn addition to talks by leading investigators, the Cancer Center’s research retreat also features the work of a new generation of cancer researchers. Graduate students, postdoctoral fellows and junior faculty members took part in the annual poster competition; the 131 presentations emphasize the breadth of studies ongoing in the Cancer Center, from cancer prevention to bioinformatics. See the award winners here. |
• 80 percent of cancer care is delivered away from the top 50 cancer centers.
• Most doctors suffer from a knowledge gap; they need accelerated genome training to understand the top molecular biomarkers and how these markers can guide patient therapy.
• Pathologists — who are a key link to alter the delivery of care — need to know not only tissue pathology but also how to test for and report the molecular drivers of cancer.
Genomics Identifies Actionable Targets
Mark Kris, M.D., an attending physician at the Memorial Sloan Kettering Cancer Center and professor at the Weill Cornell Medical College, showed how genomics and personalized care can be harnessed to improve lung cancer survival.Working with 11 cancer centers, Kris and colleagues tested 1,000 patients who had stage IV lung cancer. While tissue pathology confirmed adenocarcinoma, the cancers also underwent mutational analysis to probe for oncogenic drivers, and these findings were shared with physicians.
Two-thirds of the patients had at least one of 10 known oncogenic drivers. These drivers are “actionable targets” that helped to guide treatment choices, leading to increased median survival for these advanced cancer patients.
The French medical system, Kris noted, has provided genotyping to every lung cancer patient since 2011, at a rate of 20,000 patients a year. This equity of access to innovation does not exist in the United States, Kris said, even though the National Comprehensive Cancer Network clinical practice guidelines for non-small-cell lung cancer already list a set of molecular drivers that should be looked to to classify and guide treatment.
Needed: A New Kind of Trial
Another roadblock is the need for new ways to perform clinical trials of investigational drugs, said Donald Berry, Ph.D., professor of biostatistics at the M.D. Anderson Cancer Center and a co-founder of Berry Consultants.Berry described how the use of Bayesian biostatistics in an adaptive platform trial can lower the numbers of patients needed for the trial, while simultaneously investigating multiple drugs and targets. He focused on a current study, I-SPY2, which is investigating treatments for breast cancer. (Berry noted that UAB is one of the largest contributors of patients to the trial.)
Data obtained during trials such as I-SPY2 are used to guide changes in the studies midstream, Berry explained. The result is nimble, lean studies that yield a more dependable estimate of the chance that a particular drug will succeed in its subsequent Phase III trial. Such information is crucial, given the cost and the failure rates of conventional Phase III trials.
Predicting Patient Response With Avatars
The final outside speaker, Paul Haluska Jr., M.D., Ph.D., associate professor of oncology at the Mayo Clinic, described an “Ovarian Avatar” model to personalize ovarian cancer treatment. The avatar is created by implanting live cancer tissue from the cancer patient into a mouse within two hours of surgery.Haluska shared several definitions involved in this model:
• “Xenograft” is a tumor taken from one species and implanted in another;
• “Orthotopic” means the implant is placed in the natural body location for that type of tumor;
• “Patient-derived Xenograft” is a direct implant from the patient into the other species, without any intermediate in vitro growth or manipulation; and
• “Avatar” is thus an orthotopic, treatment-naïve, patient-derived xenograft.
Mayo implanted its first model in March 2010. Through this September, 404 models have been injected and 294 of them successfully engrafted. The avatar responses to a drug, Haluska said, appeared to mirror the patient responses to treatment with the same drug, and the avatars are being used for drug development.
The next step will be to actually use a particular patient’s avatar to direct her therapy. “It will be the first ovarian cancer with xenograft-directed therapy,” Haluska said. “The best predictor of response is response.”
Oncogenic Drivers and Racial Disparities
UAB has its own xenografts that are derived from glioblastoma multiforme tumors, said Christopher Willey, M.D., Ph.D., an associate professor in the Department of Radiation Oncology and director of the UAB Kinome Core (pronounced “k-eye-nome”). But these personal avatars have a problem — they take too much time to establish compared to the rapid and fatal course of glioblastomas. So Willey hopes instead to use “kinomic” profiles of established avatars from other patients to guide the treatment for a new patient; glioblastoma tissue removed from the new patient during surgery can quickly be kinomically profiled.Kinomics uses substrate arrays to identify which kinase enzymes — often found to be key oncogenic drivers — are active in the cancer cells. This can help select among about 30 cancer chemotherapeutic agents that target kinases.
The other UAB speaker, Phillip Buckhaults, Ph.D., associate professor in the UAB Division of Hematology and Oncology, described his search for genetic mechanisms that lead to earlier onset and higher incidence of breast and colon cancers in African-Americans, as compared to Caucasian-Americans. His trail began with the discovery of a point-mutant variant of the TP53 tumor suppressor gene in African-Americans, and it has led to the variant’s effect on the PRDM1 chromatin-silencing gene.
Translating research insights from the laboratory to the clinic is a major focus of the UAB-HudsonAlpha cancer consortium, noted Cancer Center director Edward Partridge, M.D. “We’re not at the point yet where we can routinely apply genomics information from the tumor to treatment; but we’re clearly learning, and learning at a rapid pace,” Partridge said. “The goal of the consortium is to accelerate that, and we’re excited about what it means for the care we can bring to our patients.”
— Jeff Hansen
Subscribe to:
Posts (Atom)